Melon ( Cucumis melo L.) is an important vegetable crop that has an extensive history of cultivation, and has been classified into two subspecies, C. melo ssp. agrestis and C. melo ssp. melo. Furthermore, the difference in their geographical distribution resulted in diverse characteristics between the two subspecies, shaping genomic imprinting in their genomes. Wild germplasm is an important genetic resource in crop breeding because of its high genetic diversity and resistance against diseases. However, all of the previous reported genomes were assembled based on the cultivated melon, the genome of wild and semi-wild melon types is not yet available. Therefore, assembling high-quality wild/semi-wild melon genome will provide an unprecedented opportunity for gene discovery and resistance breeding in melon.
Recently, the study of ‘The haplotype resolved T2T reference genome highlights structural variation underlying agronomic traits of melon’ was published on Horticulture Research by Melon Genetic Breeding Team from Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, collaborated with professor Tao Lin from China Agricultural University.
‘821’ (PI 313970, self-fertilized) is an accession derived from C. melo ssp. agrestis var. acidulus native to India, which possess high resistance to powdery mildew and a variety of viral diseases. Here we reported a chromosome-level T2T genome assembly for ‘821’, a semi-wild melon with two haplotypes of ~373 Mb and ~364 Mb, respectively. Comparative genome analysis discovered a significant number of structural variants (SVs) between melo and agrestis genomes, including a copy number variation located in the ToLCNDV resistance locus on chromosome 11, of which a candidate gene was identified for ToLCNDV resistance. Additionally, melon is a unique model species for studying fruit ripening because of presenting both climacteric and non-climacteric types. Genome-wide association studies detected a significant signal associated with climacteric ripening and identified one candidate gene CM_ac12g14720.1 ( CmABA2 ), encoding a cytoplasmic short chain dehydrogenase/reductase, which controls the biosynthesis of abscisic acid. This study provides valuable genetic resources for future research on resistance breeding in melon.
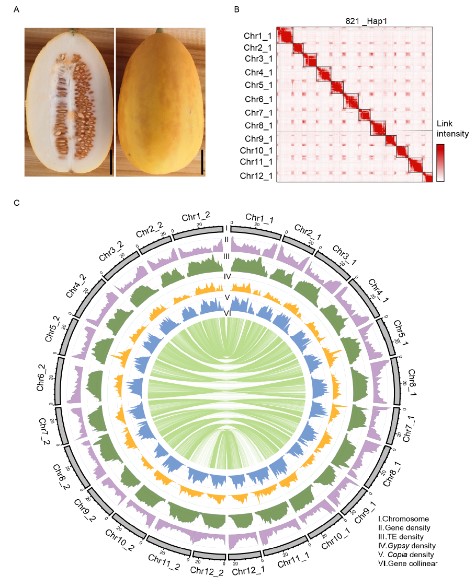
Fig.1 The phenotype, Hi-C map and genomic landscape of ‘821’
This work was supported by funding from the Agricultural Science and TechnologyInnovation Program (CAAS-ASTIP-2016-ZFRI-06), the China Agriculture Research System (CARS-25-2023-G6), the Key Research and Development Program of Hainan (ZDYF2021XDNY164), the European Research Council (ERC-NectarGland, 101095736), the 111 Project (B17043) and Henan Province Science and Technology Research Project (232102110185).
By Tang Lingli
tanglingli@caas.cn
