The new article that described the genetic basis of breeding history of peach based on genome sequencing of 480 accessions was published on Genome Biologyat February 22, 2019. This study providesnew insights into the genetic footprints for peach during domestication and improvement and how human selection shapes the genomes, which is helpful for future genomic selection and molecular breeding.This study was mainly finished by Prof. Lirong Wang’s lab from Zhengzhou Fruit Research Institute, CAAS, cooperated with Prof. Wenwu Guo from Huazhong Agricultural University and Prof. Zhangjun Fei from Boyce Thompson Institute for Plant Research in Cornell University.
Peach originates from China with abundant genetic resources in China. We sequence a total of 480 wild and cultivated peach accessions from around the world which captured more than 95% genetic diversity of the collection of 1,410 accession in National Peach Germplasm Repository of China (Zhengzhou). Based on sequencing data, we construct thehitherto biggest variation map for peach. Using the variation map, we perform the genome-wide association study (GWAS) on 11 importantly agronomic traits and mine several novel associated loci, which provide candidate for marker development and molecular breeding. We identify a total of 142 and 104 selective sweeps during domestication and improvement, respectively. Most of selective sweeps harbor the loci related to agronomic traits (See following figure). By focusing on a set of quantitative trait loci (QTLs), we provide evidence supporting thatdistinct phases of domestication and improvement have led to an increase in fruit size and taste and extended itsgeographic distribution. Fruit size was predominantly selected during domestication, and selection for large fruitshas led to the loss of genetic diversity in several fruit weight QTLs. In contrast, fruit taste-related QTLs weresuccessively selected for by domestication andimprovement, with more QTLs selected for during improvement. Novel loci for chilling requirement and candidate loci for genes that contributed to the adaption tolow-chill regions were identified. Moreover, a PCR-based marker for early selection of low chilling requirement is developed. Furthermore, the genomic bases of divergent selection for fruit texture and localbreeding for different flavors between Asian and European/North American cultivars were also determined.
The article is a newly important progress of the 1,000 Peach Genome Project designed and implemented by Wang’s lab. Her lab has previously published two articles focusing on peach genomics on Genome Biology and Nature Communications at 2014 and 2016, respectively. This article comprehensively elucidates the genomic footprints of artificial selection during domestication and improvement, which provides important resources for future molecular design breeding. Further works are underway.
The first author and corresponding author are PhD candidate Yong Li and Prof. Lirong Wang, respectively. This study was supported by the grants from the National Natural ScienceFoundation of China (31572094), the Agricultural Science and TechnologyInnovation Program (CAAS-ASTIP-2018-ZFRI-01), Crop Germplasm ResourcesConservation Project (2016NWB041).
The URL for this article should be:
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1648-9
By Yong Li
jay20075267@163.com
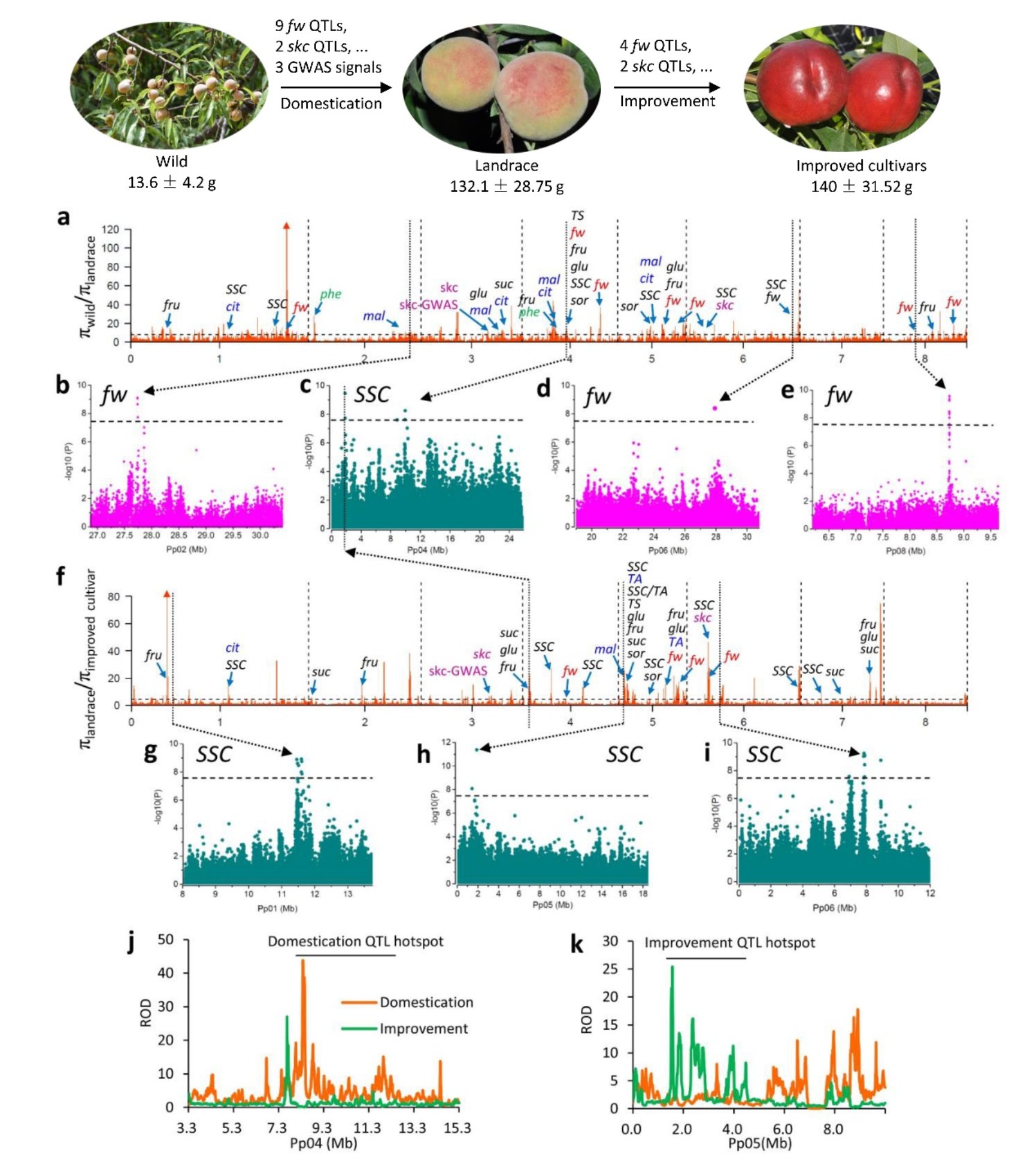
Figure Genome-wide detection and functional annotations of selection sweeps during domestication and improvement.
