Kiwifruit resources and breeding innovation team from the Zhengzhou Fruit Research Institute published an article “Development of a 135K SNP genotyping array for Actinidia argutaand its applications for genetic mapping and QTL analysis in kiwifruit” in the Plant Biotechnology Journal.
Kiwifruit (Actinidia spp) is a woody, perennial and dioecious vine. In this genus, there aremultiple ploidy levels but the main cultivated cultivars are polyploid. Despite the availability ofmany genomic resources in kiwifruit, SNP genotyping is still a challenge given these differentlevels of polyploidy. In this study, we developed a high-density SNP genotyping array to facilitate genetic studies and breeding applications in kiwifruit.We screened a germplasm collection to identify SNPs across Actinidia species, filtered and selected 135K SNPs for the array.The array was used to genotype a tetraploid Actinidia argutaF1 population.An integrated linkage map wasconstructedcovering 3060.9 cM across 29 linkage groups, showing a high degree of collinearity between the A. arguta genetic map and the A. chinensis Red5 reference genome. QTL mapping for the sex locus was performed that led to identification of a novel QTL on Linkage Group 3. Finally, the study presented evidence of tetrasomic inheritance with partial preferential pairing in A. arguta.
The study was supported by the National Key Research and Development Project of China(2019YFD1000802); the National Natural Science Foundation of China (32001995) and the Science and Technology Innovation Project of CAAS(CAAS-ASTIP-2015-ZFRI).
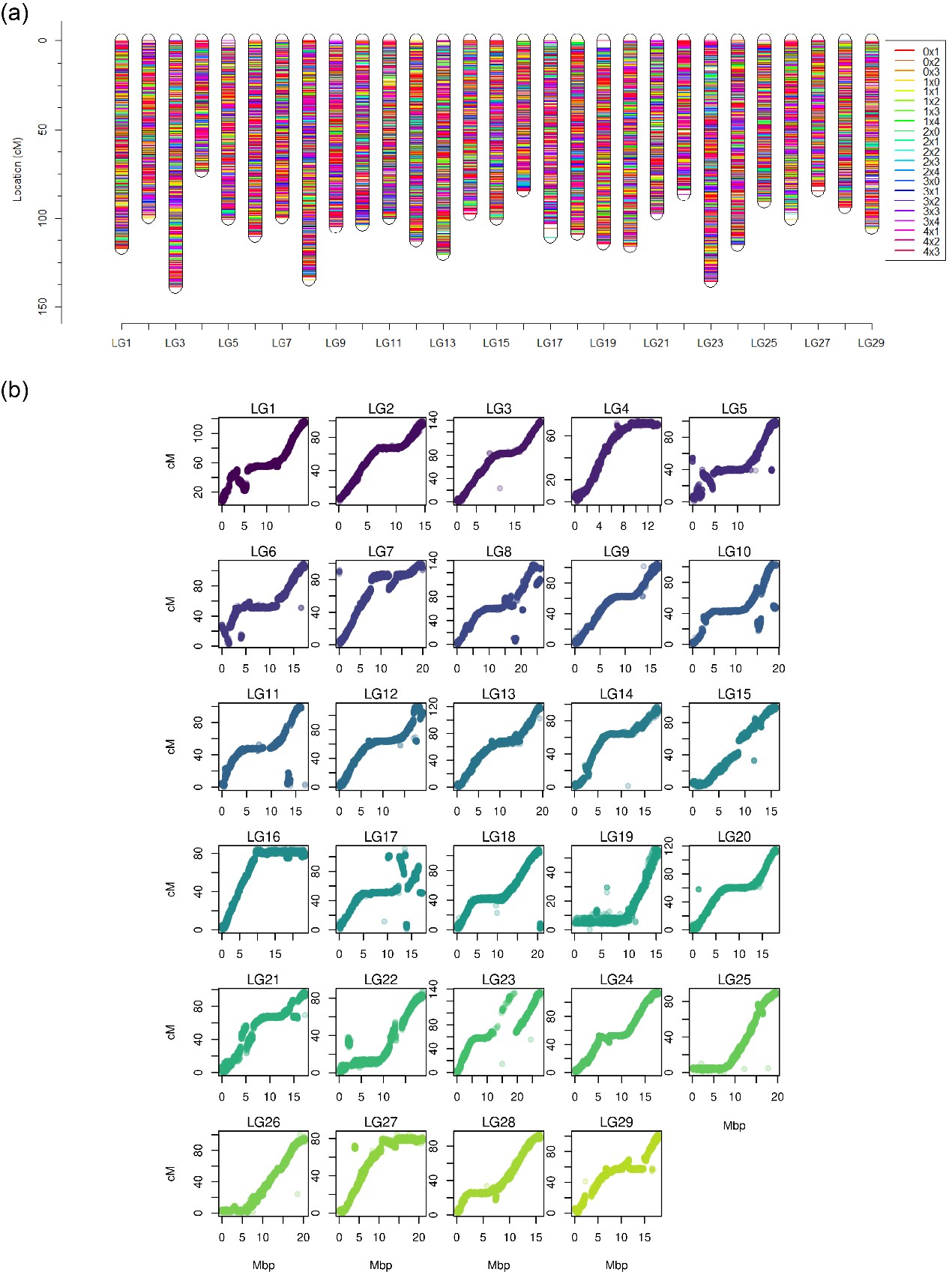
The genetic map of tetraploid Actindia argutaand comparison of the genetic map with thekiwifruit genome
