Researchers from the Zhengzhou Fruit Research Institute (ZFRI) of the Chinese Academy of Agricultural Sciences have published a study titled “Combination of 3D chromatin architecture and omics analysis provides insight into anthocyanin regulation in Actinidia arguta” in Horticulture Research. The research is the first to elucidate the 3D genomic basis underlying the differential formation of fruit skin color in this species.
The three-dimensional (3D) genome plays a crucial role in regulating gene expression and shaping biological traits. In eukaryotes, higher-order chromatin structure is organized into distinct functional units. While the skin color of A. arguta is primarily either red or green, it has been unclear whether 3D genome architecture is involved in its regulation.
Building on the the first high-quality reference genome of a whole red A. arguta, this study employed high-through chromosome conformation capture (Hi-C) to generate 3D genome data for the red-skinned ‘ZHB’ and green-skinned ‘ZLB’. The team compared differences at multiple levels, including compartments, TADs and loops. By integrating comparative genomics, transcriptome, and ATAC-seq analysis, they discovered that a 346 bp structural variation in the promoter of the calmodulin-binding protein gene AaCBP60B-like affacts fruit skin coloration by disrupting TADs, rebuilding loops, and altering promoter activity. The gene’s role in anthocyanin regulation was further confirmed through exogenous calcium chloride treatments, transient overexpression and gene silencing experiments.
In summary, this study construncted the first 3D genome map of A. arguta and identified a key gene, AaCBP60B-like, along with a critical genetic variation that regulate fruit skin color. These findings provide novel insights into the epigenetic regulation of kiwifruit coloration and present a new target for gene-editing-mediated color breeding, holding significant theoretical and practical value.
Associate Research Fellow Yukuo Li, Master’s student Zhe Song, and Xu Zhan are co-first authors of the study. Research Fellows Xiujuan Qi and Jinbao Fang are co-corresponding authors. Master’s student Xiaohan Li and PhD student Lingshuai Ye also contributed to the research. This work was supported by grants from the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-ZFRI-03-202502), the National Key Research and Development Program of China (2022YFD1200503), the National Natural Science Foundation of China (32202435), the China Agriculture Research System of MOF and MARA (CARS-26), the Agricultural Science and Technology Innovation Program, and the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2023-ZFRI-03).
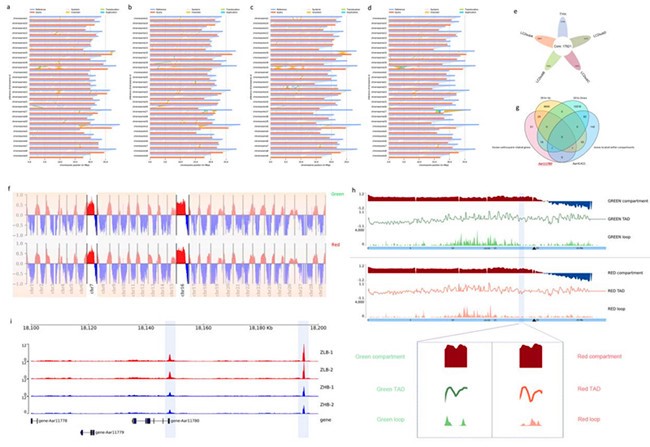
Screening key candidate gene through multi-omics integrative analysis
Article link:https://doi.org/10.1093/hr/uhaf183
By Li Yukuo
liyukuo@caas.cn
