The new article that assembled the soft-seeded pomegranate ‘Tunisia’ genome was published on Plant Biotechnology Journal at September 24, 2019. This study provides new insights into the genetic divergence between soft- and hard-seeded cultivars in pomegranate, which is helpful for future molecular improvement.
Pomegranate ( Punica granatum L.) is an important edible fruit tree species native to central Asia. The fruit has gained widespread popularity because of its functional and nutraceutical properties. The properties of pomegranate juices, seeds, and extracts are potentially beneficial for treating cardiovascular disease, diabetes, and prostate cancer. Traditional hard-seeded pomegranate varieties are well adapted to the cold environment, whereas the soft-seeded varieties have become popular with consumers as their fruits are easily swallowed. Additionally, soft-seeded varieties fetch high price in the market as compared to hard seeded. To date, ‘Tunisia’ has been the only one commercial variety introduced from Tunisia since 1986. The problem of single variety and depression seriously restricts the development of pomegranate industry. Complete and highly accurate reference genomes and gene annotations are indispensable for basic biological research and trait improvement of pomegranate.
In this study, we integrated single-molecule sequencing and high-throughput chromosome conformation capture techniques to produce a high-quality and long-range contiguity chromosome-scale genome assembly of the soft-seeded pomegranate cultivar‘Tunisia’. The genome covers 320.31 Mb (contig N50 = 4.49 Mb) and includes 33,594 protein-coding genes. 97.76% (313.13 Mb) of the assembly was anchored on eight chromosomes. The length of contig N50 for the new‘Tunisia’reference genome was 46-fold greater than that for the recently published‘Taishanhong’genomes.
Comparative genomic analyses revealed many genetic differences between soft- and hard-seeded pomegranate varieties. Re-sequenced 26 pomegranate varieties revealed a set of selective loci. An exceptionally large selective region (26.2 Mb) was identified on chromosome 1. Gene annotation indicate that genomic variations and selective genes may have contributed to the genetic divergence between soft- and hard-seeded pomegranate varieties.
The corresponding author and first author Prof. Shangyin Cao and PhD Xiang Luo, respectively. This project was funded by the Key Project of the National Science and Technology Basic Work of China (2012FY110100) and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2015-ZFRI).
The URL for this article should be: https://onlinelibrary.wiley.com/doi/abs/10.1111/pbi.13260
By Xiang Luo
Email: tjaulx@126.com
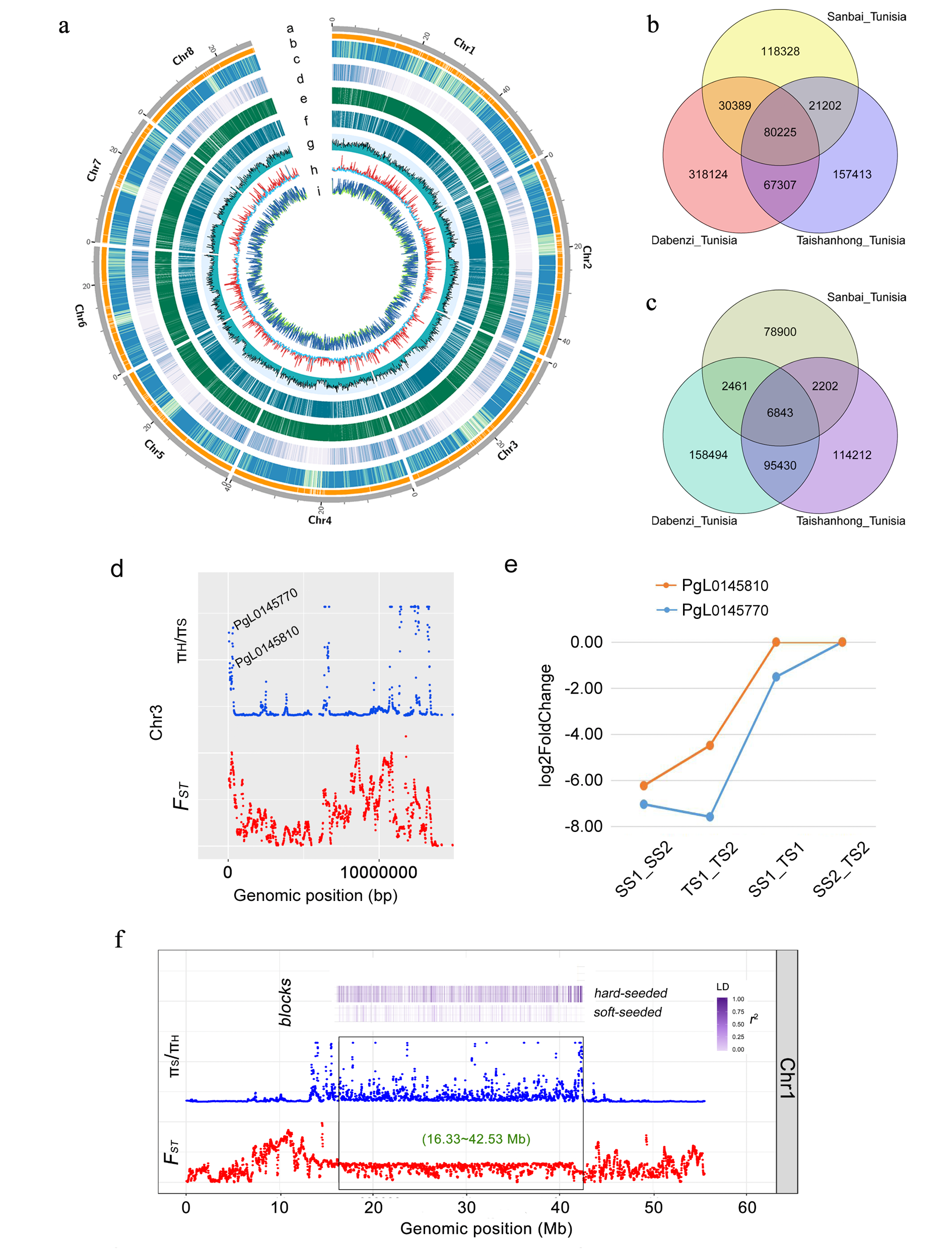
Figure1 Genome assembly and genetic factors associated to the genetic divergence between soft- and hard-seeded cultivars
 |
 |
Figure 2 ‘Tunisia’ cultivar
